This vignette shows how to generate a Full Factorial
Design using both the FielDHub Shiny App and the scripting
function full_factorial() from the FielDHub
package.
1. Using the FielDHub Shiny App
To launch the app you need to run either
FielDHub::run_app()or
Once the app is running, go to Other Designs > Full Factorial Designs
Then, follow the following steps where we show how to generate this
kind of design by an example with a set of 3 treatments with levels
3, 3, 2 each. We will run this experiment 3 times.
Inputs
-
Import entries’ list? Choose whether to import a
list with entry numbers and names for genotypes or treatments.
If the selection is
No, that means the app is going to generate synthetic data for entries and names of the treatment based on the user inputs.If the selection is
Yes, the entries list must fulfill a specific format and must be a.csvfile. The file must have two columns:FACTORSandLEVEL. Containing a list of unique names that identify each treatment and level. Duplicate values are not allowed, all entries must be unique. In the following table, we show an example of the entries list format. This example has an entry list with three treatments/factors, and 3, 3 and 2 levels each.
| FACTOR | LEVEL |
|---|---|
| A | a0 |
| A | a1 |
| A | a2 |
| B | b0 |
| B | b1 |
| B | b2 |
| C | c0 |
| C | c1 |
Choose whether to use the factorial design in a RCBD or CRD with the Select a Factorial Design Type box. Set it to
RCBD.Set the number of entries for each factor in a comma separated list in the Input # of Entries for Each Factor box. We want our example experiment to have 3 factors with 3, 3, and 2 levels respectively, so enter
3, 3, 2in the box.Set the number of replications of the squares with the Input # of Full Reps box. Set it to
3.Enter the number of locations in Input # of Locations. Set it to
1.Enter the starting plot number in the Starting Plot Number box. If the experiment has multiple locations, you must enter a comma separated list of numbers the length of the number of locations for the input to be valid. In this case, set it to
101.Optionally, you may enter a name for the location of the experiment in the Input Location box.
Select
serpentineorcartesianin the Plot Order Layout. For this example we will use the defaultserpentinelayout.As with all the designs, we can set a random seed in the box labeled random seed. In this example, we will set it to
1239.Once we have entered the information for our experiment on the left side panel, click the Run! button to run the design.
Outputs
After you run a full factorial design in FielDHub, there are several ways to display the information contained in the field book.
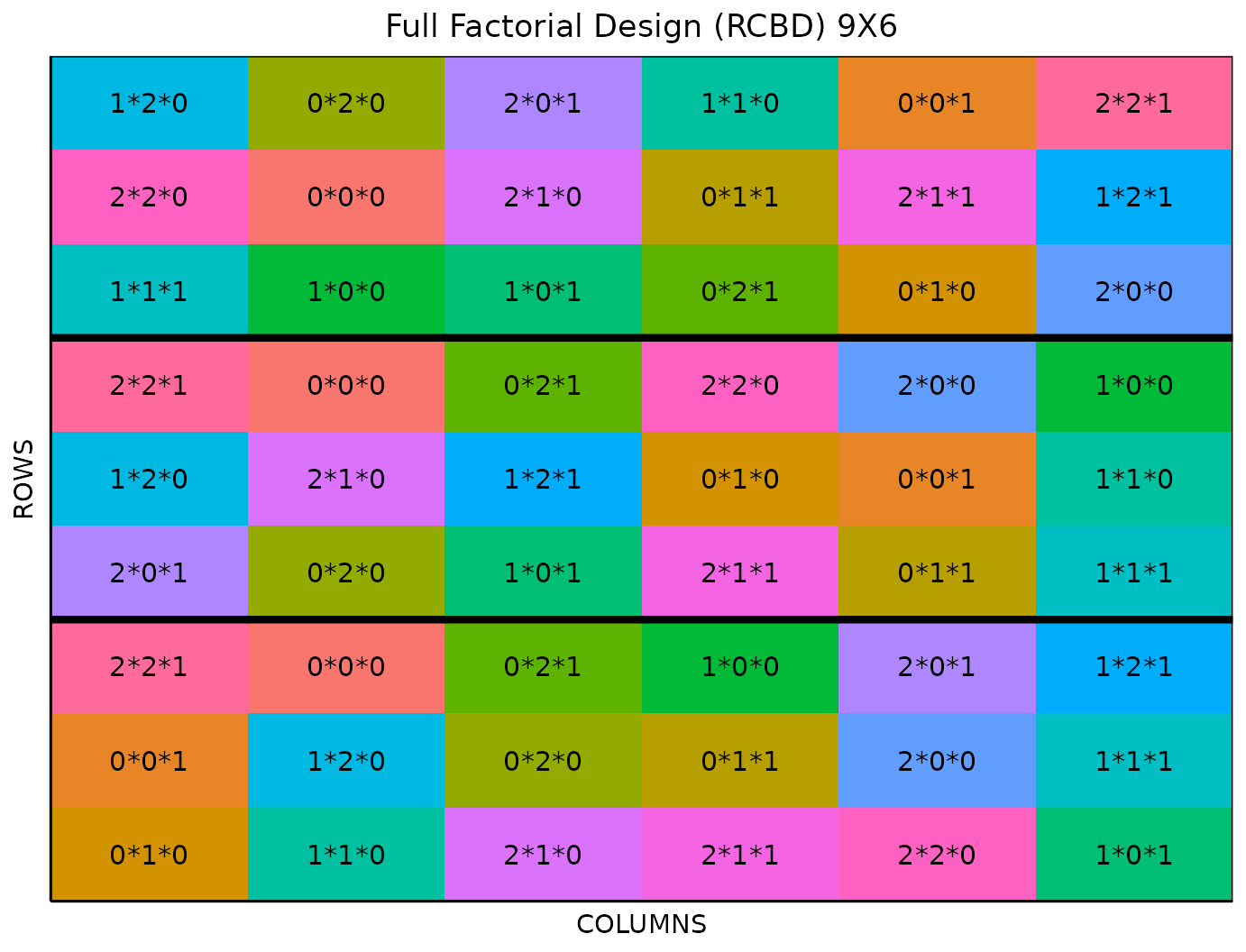
Field Layout
When you first click the run button on a full factorial design,
FielDHub displays the Field Layout tab, which shows the entries and
their arrangement in the field. In the box below the display, you can
change the layout of the field or change the location displayed. You can
also display a heatmap over the field by changing Type of
Plot to Heatmap. To view a heatmap, you must first
simulate an experiment over the described field with the
Simulate! button. A pop-up window will appear where you
can enter what variable you want to simulate along with minimum and
maximum values.
Field Book
The Field Book displays all the information on the experimental design in a table format. It contains the specific plot number and the row and column address of each entry, as well as the corresponding treatment on that plot. This table is searchable, and we can filter the data in relevant columns. If we have simulated data for a heatmap, an additional column for that variable appears in the Field Book.
2. Using the FielDHub function:
full_factorial()
You can run the same design with a function in the FielDHub package,
full_factorial().
First, you need to load the FielDHub package typing,
Then, you can enter the information describing the above design like this:
factorial <- full_factorial(
setfactors = c(3,3,2),
reps = 3,
l = 1,
type = 2,
plotNumber = 101,
planter = "serpentine",
locationNames = "FARGO",
seed = 1239
)Details on the inputs entered in full_factorial()
above
The description for the inputs that we used to generate the design,
-
setfactors = c(3,3,2)are the levels of each factor. -
reps = 3is the number of replications for each treatment. -
l = 1is the number of locations. -
type = 2means CRD or RCBD, 1 or 2 respectively. -
plotNumber = 101is the starting plot number. -
planter = "serpentine"is the order layout. -
locationNames = "FARGO"is an optional name for each location. -
seed = 1239is the random seed to replicate identical randomizations.
Print factorial object
print(factorial)Full Factorial Design
Information on the design parameters:
List of 9
$ factors : chr [1:3] "A" "B" "C"
$ levels : int [1:8] 0 1 2 0 1 2 0 1
$ runs : int 18
$ all_treatments :'data.frame': 18 obs. of 3 variables:
..$ A: int [1:18] 0 1 2 0 1 2 0 1 2 0 ...
..$ B: int [1:18] 0 0 0 1 1 1 2 2 2 0 ...
..$ C: int [1:18] 0 0 0 0 0 0 0 0 0 1 ...
$ reps : num 3
$ locations : num 1
$ location_names : chr "FARGO"
$ kind : chr "RCBD"
$ levels_each_factor: num [1:3] 3 3 2
10 First observations of the data frame with the full_factorial field book:
ID LOCATION PLOT REP FACTOR_A FACTOR_B FACTOR_C TRT_COMB
1 1 FARGO 101 1 0 1 0 0*1*0
2 2 FARGO 102 1 1 1 0 1*1*0
3 3 FARGO 103 1 2 1 0 2*1*0
4 4 FARGO 104 1 2 1 1 2*1*1
5 5 FARGO 105 1 2 2 0 2*2*0
6 6 FARGO 106 1 1 0 1 1*0*1
7 7 FARGO 107 1 0 0 1 0*0*1
8 8 FARGO 108 1 1 2 0 1*2*0
9 9 FARGO 109 1 0 2 0 0*2*0
10 10 FARGO 110 1 0 1 1 0*1*1Access to factorial object
The full_factorial() function returns a list consisting
of all the information displayed in the output tabs in the FielDHub app:
design information, plot layout, plot numbering, entries list, and field
book. These are accessible by the $ operator,
i.e. factorial$layoutRandom or
factorial$fieldBook.
factorial$fieldBook is a list containing information
about every plot in the field, with information about the location of
the plot and the treatment in each plot. As seen in the output below,
the field book has columns for ID, LOCATION,
PLOT, REP, and TRT_COMB, and
columns for each factor individually.
field_book <- factorial$fieldBook
head(factorial$fieldBook, 10) ID LOCATION PLOT REP FACTOR_A FACTOR_B FACTOR_C TRT_COMB
1 1 FARGO 101 1 0 1 0 0*1*0
2 2 FARGO 102 1 1 1 0 1*1*0
3 3 FARGO 103 1 2 1 0 2*1*0
4 4 FARGO 104 1 2 1 1 2*1*1
5 5 FARGO 105 1 2 2 0 2*2*0
6 6 FARGO 106 1 1 0 1 1*0*1
7 7 FARGO 107 1 0 0 1 0*0*1
8 8 FARGO 108 1 1 2 0 1*2*0
9 9 FARGO 109 1 0 2 0 0*2*0
10 10 FARGO 110 1 0 1 1 0*1*1